
Biotechnological
Communication
Biosci. Biotech. Res. Comm. 11(3): 505-511 (2018)
Biodegradation of cotton seed soapstocks by novel
indigenous
Bacillus species
Gayatriben B. Patel and Kamlesh R. Shah
Department of Biotechnology,Pramukh Swami Science and H.D. Patel Arts College, Kadi, India
ABSTRACT
Soapstocks is a value-added by-product separated from vegetable oil re ning operations. Bacillus sp. is a good
enzyme producer. The present research paper focused at isolation, screening and identi cation of Bacillus sp. from
soapstock samples. Cotton seed soapstock samples used in the study were 7% oil rich gelatinous dark brown chemi-
cal compound, which were enriched, serially diluted and spread on tributyrin agar plates, to isolate lipase positive
cultures. Qualitative analysis of lipase producing microorganisms was done by plate assay on tributyrin agar plate
and zone of hydrolysis measured. Bacillus sp. were further screened for cellulase and protease production by plate
assay. Two cultures were identi ed on the basis of molecular and biochemical characteristics as Bacillus licheniformis
(3B) and Bacillus pumilus (18B). Among selected Bacillus cultures Bacillus licheniformis and Bacillus pumilusgave-
good zone of lipase and cellulasehydrolysis. Bacillus pumiluswas highly protease producing organism. Quantitative
analysis of Lipase production activity measured spectrophotometrically using p-nitrophenyl palmitate (p-NPP) as
substrate. Bacillus licheniformis showed 1.72 U/ml lipase productions whereas Bacillus pumilus(18B) has 2.59 U/ml.
Phylogenetic trees showed similarity with other highly similar species.
KEY WORDS:
BACILLUS LICHENIFORMIS
,
BACILLUS PUMILUS,
LIPASE, P-NPP, PHYLOGENETIC TREES
505
ARTICLE INFORMATION:
*Corresponding Author: hisonu2408@yahoo.co.in
Received 12
th
July, 2018
Accepted after revision 18
th
Sep, 2018
BBRC Print ISSN: 0974-6455
Online ISSN: 2321-4007 CODEN: USA BBRCBA
Thomson Reuters ISI ESC / Clarivate Analytics USA and
Crossref Indexed Journal
NAAS Journal Score 2018: 4.31 SJIF 2017: 4.196
© A Society of Science and Nature Publication, Bhopal India
2018. All rights reserved.
Online Contents Available at: http//www.bbrc.in/
DOI: 10.21786/bbrc/11.3/21
INTRODUCTION
Industrial wastes, vegetable oil processing factories,
soil contaminated with oil etc contain oily environment
which provides a good environments for lipase producing
microorganisms (Vandana et al., 2014). Soapstocks is a
gelatinous dark brown undesirable chemical compound
product from vegetable oil re ning operations (King
et al., 1998). Crude oil contamination in the environment
has lots of hazard and so remediation of crude oil creates
area of interest for research (Guru et al., 2013). Microbes
secrete various enzymes among them lipase which helps

506 BIODEGRADATION OF COTTON SEED SOAPSTOCKS BY NOVEL INDIGENOUS
BACILLUS SP.
BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS
Gayatriben B. Patel and Kamlesh R. Shah
in degradation of oil (Veerabagu et al., 2014). Research
in bacterial lipases is of great demand now because of
value added potential industrial application (Sirisha
et al., 2010). Industries are seeking lipase producing
strains of bacteria which contain excellent properties
using cost effective methods on large scale production
(Bharathi et al., 2018).
Lipase (triacyl glycerol acylhydrolases, EC 3.1.1.3)
catalyzes the hydrolysis of triacyl glycerol to glycerol
and long chain fatty acids at oil water interface (Pualsa
et al., 2013). Research can be done toward lipases from
plant and animal origin but lipases from microbial ori-
gin are receiving much attention with the rapid develop-
ment of enzyme technology. Lipase act as biocatalysts
constitute the signi cant important role for biotechno-
logical applications (Hasan et al., 2006, Saxena et al.,
1999). Microbial lipases constitute much application
such as in the detergent industry, food industry, paper
and pulp industry, organic synthesis, bioconversion in
aqueous media, resolution of racemic acids and alco-
hols, regioselectiveacylations, ester synthesis, oleochem-
ical industry and lipases in medical application (Sharma
et al., 2001, Verma et al., 2012, Mauti et al., 2016, Sar-
aswat et al., 2017)
This study was conducted to isolate lipase producing
bacteria which were screened on tributyrin agar plates.
They were further analyzed for cellulase and protease
production by plate assay. The bacterial genus Bacil-
lus were identi ed on the basis of biochemical tests
and molecular 16s r DNA Partial Gene sequencing ana-
lyzes. Quantitative analysis of lipase production was
done spectrophotometrically using p-NPP as substrate.
Further study will conducted on enzymatic degrada-
tion.
MATERIAL AND METHODS
Soapstock samples were collected from two different
cotton oil re nery industries nearby Kadi (North Guja-
rat), India. At the starting season of cotton (November),
Soapstock samples were collected from the owing
stock at Washer discharge end of the pipe in a sterile
and air tight container. B/H (Bushnell–Haas) medium
was selected for enrichment of cotton seed oil soap-
stocks for microbial growth (Guru et al., 2013).10 gram
of cotton seed oil soapstock samples were added to 100
ml of B/H mediums and incubated at 37C in static con-
dition for 5 days. From each sample, 1ml of enriched
samples were transferred to the 100 ml of Tributyrin
broth medium incubated at 37 C, in shaking condition
at 100rpm for 48 hours. Enrichment was performed over
a 7 days of incubation.Enriched Soapstock samples were
serially diluted
.
Diluted samples were spread on to Tribu-
tyrin agar medium for isolation of Bacteria .TBA Plates
were incubated at 37C for 2days. Isolated colonies
were puri ed on same medium by streak plate method.
Pure cultures isolate were preserved at low temperature
in Nutrient agar slants for screening and further use.
Lipase-producing strains were screened by qualitative
plate assay according to Lokre et al., 2014. Isolates were
spot inoculated on tributyrin agar plates and incubated
at 37°C for 2 days. Zone of clearance was observed due
to hydrolysis of tributyrin by lipase enzyme.
Cellulase and Protease activity were done by qualita-
tive agar plate assay in nutrient agar media containing
respective substrates. Culture was spot inoculated and
incubated at 37 °C for 2 days. Check for the zone of
clearance around the colonies due to utilization of the
particular substrate.
Culture was grown in medium containing 1% car-
boxy methyl cellulose (Dabhi et al., 2014). After incu-
bation the CMC plates were ooded with 0.1 % congo
red staining, after 5 min stain was discarded and the
plates were distained by 1M NaCl solution with con-
tinuous stirring for 15-20 min. The clear zone around
colonies indicated cellulose hydrolysis.Protease activ-
ity was checked in medium containing 1% skim milk
as substrate (Prabavathi et al., 2012). Spot inoculated
cultures were incubated at 37 °C for 2 days and observed
for clearance zone around colonies.
Selected Bacterial cultures that show Positive lipase
production in plate assay, which were subjected fur-
ther for Quantitative estimation. 2 days old bacterial
cultures grown on TBA medium were used for inocula-
tion. One loopfull culture was inoculated into 100 ml
of inoculum medium containing: peptone 0.5%, Yeast
extract 0.5%, NaCl 0.5% and cotton seed Oil 1%. Cul-
tures were incubated at 37C and 100 rpm for 4hrs. 5%
inoculum medium was further inoculated into 100 ml
of same medium (as mentioned above) for lipase pro-
duction and incubated at 37C and 100 rpm for 5 days.
Enzyme assay was performed according to the method
by Winkler et al., 1997 with some modi cation. The cul-
ture ltrate (production medium) was removed at 24 hr
interval from each ask & centrifuged at 10,000 rpm for
10 min at 4C. Supernatant was used for enzyme assay.
Lipase activity was determined by a spectrophotometric
assay using p-nitrophenyl palmitate (pNPP) as substrate.
P-NPP was hydrolysed by lipase to give p-NP which
gave yellow color, absorbance of which was measured
spectrophotometrically at 410 nm against enzyme free
blank. Statistical Analysis were done in Microsoft word
excel data analysis of lipase production.
The isolates showing maximum zone of clearance
were selected for further analysis. Morphological and
biochemical characteristics of the isolates were stud-
ied for the identi cation of the potent Bacterial isolate.
Molecular characterization of potent Bacterial strains
BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS BIODEGRADATION OF COTTON SEED SOAPSTOCKS BY NOVEL INDIGENOUS
BACILLUS SP.
507
Gayatriben B. Patel and Kamlesh R. Shah
was done by 16s rDNA partial Gene sequencing analy-
sis. It was carried out at Biogene department of GSBTM,
Gandhinagar.
The bacterial isolates were identi ed on the basis of
their morphological characteristics (like cell shape, Gram
staining, spore staining and motility) and biochemical
tests viz. According to Cappuccino et al.,1996 biochemi-
cal test were done like Voges Proskaurtes test, Citrate
utilization, Gelatin hydrolysis, Nitrate reduction, Orni-
thine decarboxylase, Lysine decarboxylase, Catalase
test and hydrolysis, Indole test, Starch hydrolysis, H2S
production, and Gas production from glucose. The uti-
lization of different sugars was studied using bacterial
identi cation kit from HiMedia.
MOLECULAR IDENTIFICATION USING 16S
RDNA SEQUENCING
Two bacterial isolates (3B and 18B) were identi ed using
16S rDNA sequencing. DNA was isolated from these
bacterial isolates and its quality was evaluated on 1.2%
agarose gel. The 16S rDNA gene was ampli ed by PCR
from the above isolated DNA and the PCR amplicon was
puri ed to remove contaminants. 16S rDNA gene was
generated from forward and reverse sequence data using
aligner software. The 16S rDNA gene sequence was used
to carry out BLAST with the nr database of NCBI gene
bank database. Based on maximum identity score rst
ten sequences were selected and aligned using multiple
alignment software program Clustal W. Distance matrix
was generated using RDP database and the phylogenetic
tree was constructed using MEGA 4.
RESULTS AND DISCUSSION
SCREENING AND ISOLATION OF LIPOLYTIC
BACTERIA
From enriched Soapstock samples, total 49 pure cultures
were isolated. Among them 30 cultures were bacterial
isolates. All 30 bacterial isolates were lipase positive, 6
bacterial cultures were protease producers and 10 bacte-
rial cultures were cellulase producers. Best two highly
positive cultures were selected on the basis of qualita-
tive analysis of lipase, cellulase and protease by plate
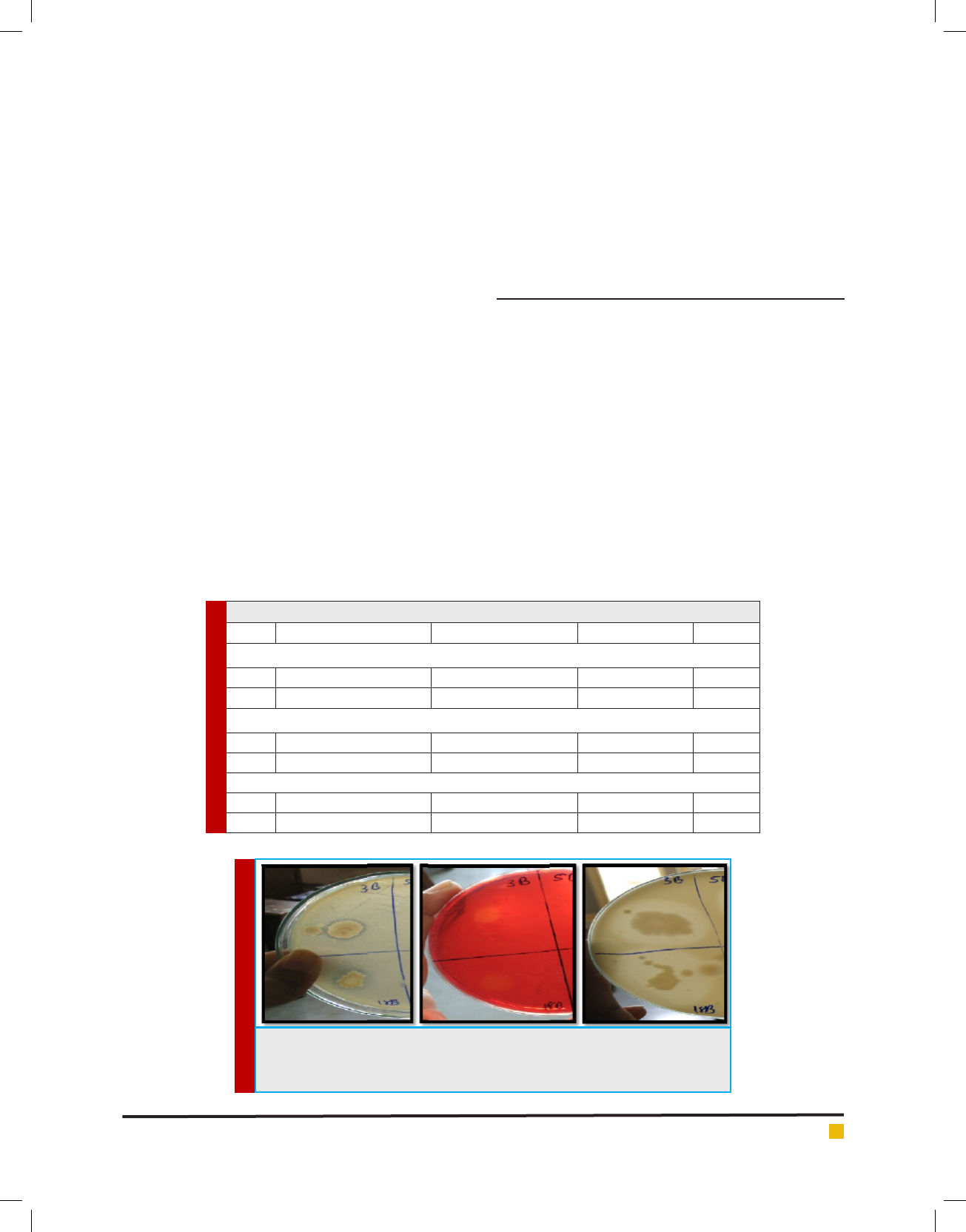
assay as shown in Table 1 and Fig 1. In 2014 Ali et al,
reported that the lipolytic bacterial Spp. isolated from oil
contaminated soil were dominantly from genus Bacillus
and Psudomonas with 23% percentage of occurrence of
Bacillus spp. among different bacteria in samples, fol-
Table.1 Measures of Clear zone diameter to colony diameter ratio of bacterial isolates.
Sr. no Bacterial Isolate Clear Zone Diameter Colony Diameter Ratio
Lipase activity
1 Bacillus licheniformis (3B) 20 mm 15 mm 1.33 mm
2 Bacillus pumilus (18B) 22 mm 16 mm 1.37 mm
Cellulase activity
1 Bacillus licheniformis (3B) 9 mm 5 mm 1.80 mm
2 Bacillus pumilus (18B) 9 mm 5 mm 1.80 mm
Protease activity
1 Bacillus licheniformis (3B) 30 mm 16 mm 1.87 mm
2 Bacillus pumilus (18B) 31 mm 10 mm 3.10 mm
FIGURE 1. A. Lipase positive cultures on 1% tributyrin agar plate. B. Cellulase
positive cultures on 1% CMC agar plate. C. Protease positive cultures on 1% Skim
milk agar plate
Gayatriben B. Patel and Kamlesh R. Shah
508 BIODEGRADATION OF COTTON SEED SOAPSTOCKS BY NOVEL INDIGENOUS
BACILLUS SP.
BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS
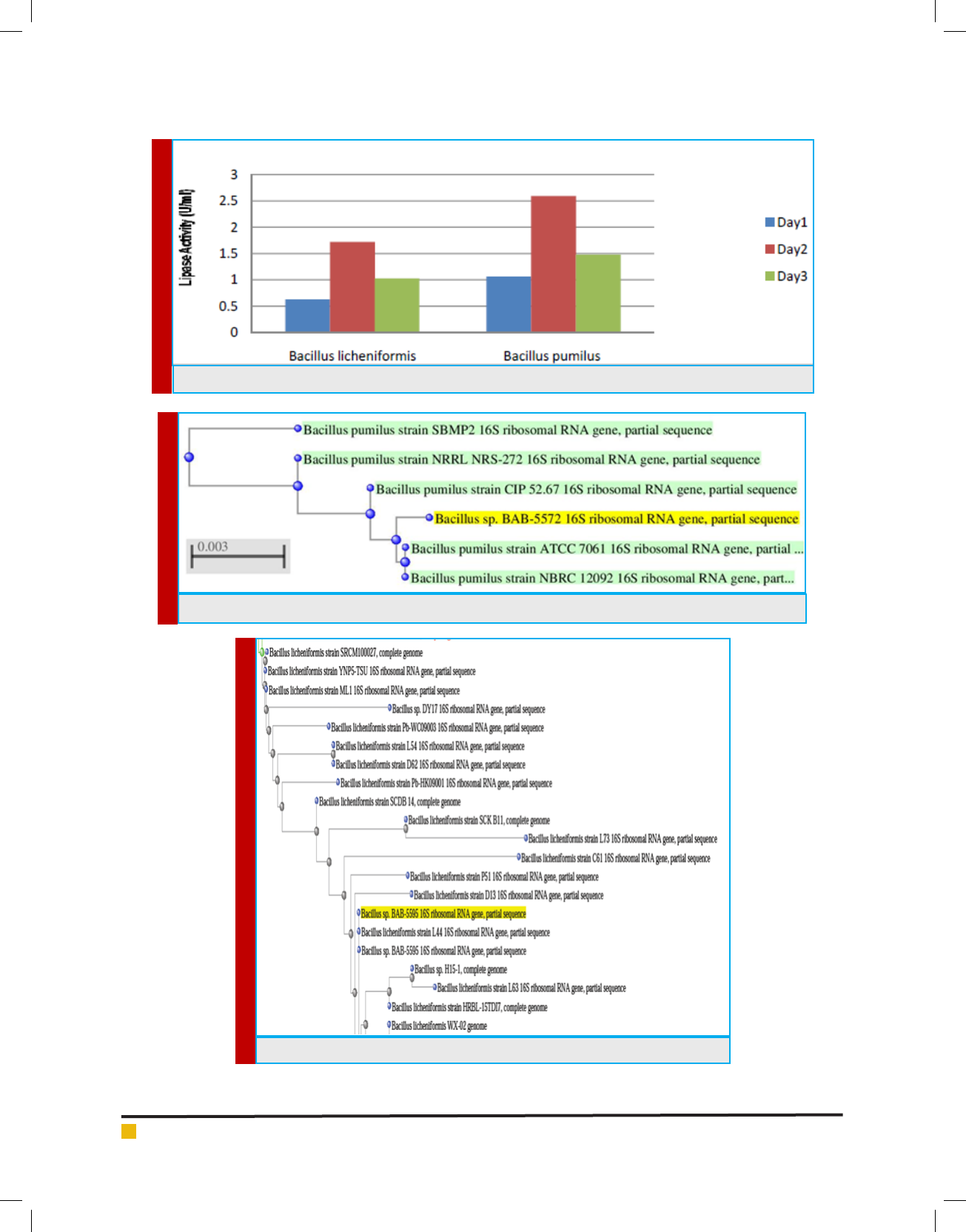
FIGURE 2. Lipase production of bacterial isolates in U/ml/min.
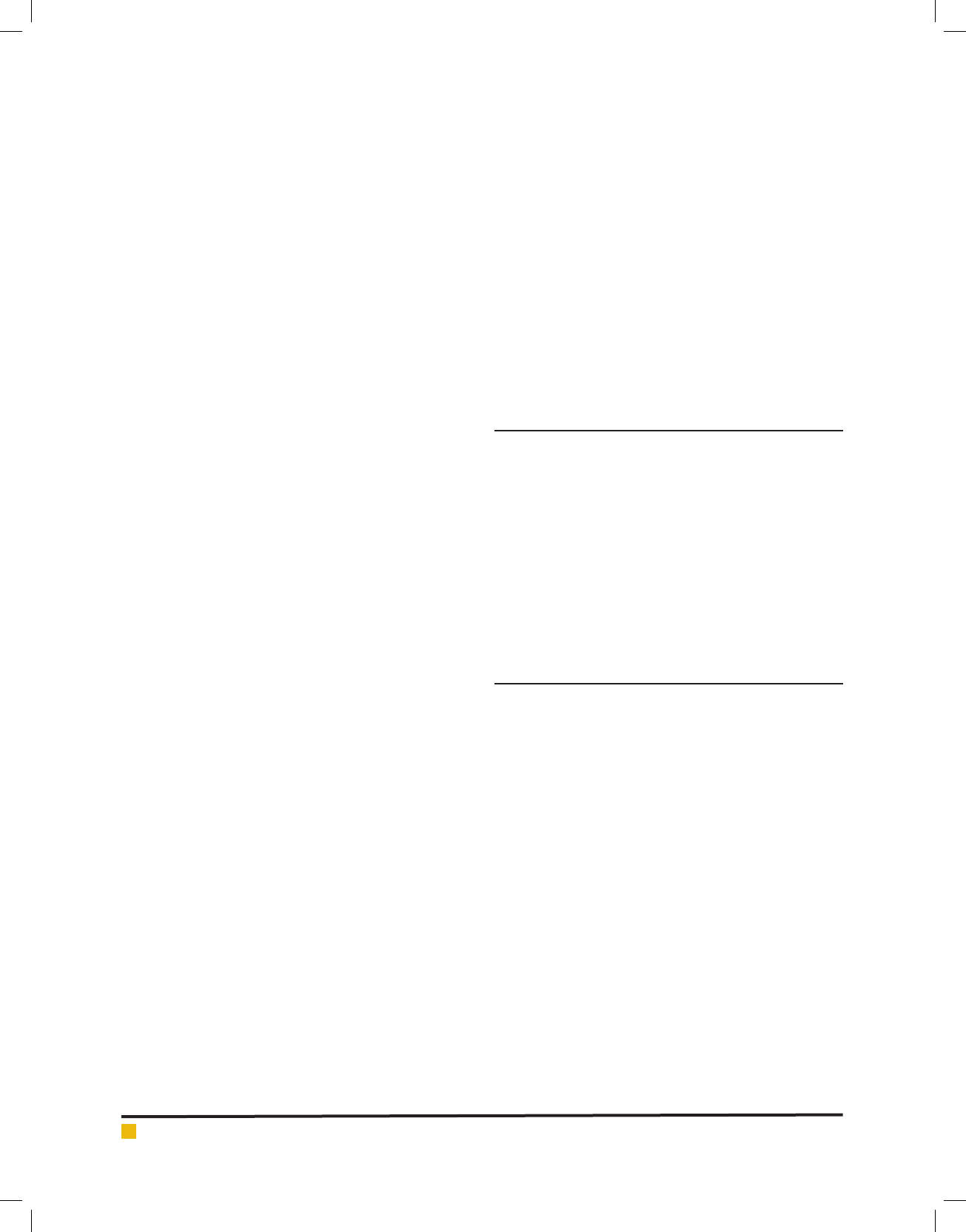
FIGURE 3. Phylogenetic tree of 18B Bacillus pumilus
FIGURE 4. Phylogenetic tree of 18B Bacillus licheniformis.

Gayatriben B. Patel and Kamlesh R. Shah
BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS BIODEGRADATION OF COTTON SEED SOAPSTOCKS BY NOVEL INDIGENOUS
BACILLUS SP.
509
Table 3. Morphological and Biochemical test of isolated bacteria
Sr. no Biochemical test Bacillus licheniformis (3B) Bacillus pumilus(18B)
1 Gram’s staining Gram positive Gram positive
2 Motility + +
3 Endospore - -
4 Methyl red + +
5 VogesProskauer’s + +
6 Citrate Utiliation + +
7 Indole + +
8 Glucuronidase + +
9 Nitrate reduction + +
10 PYR + +
11 ONPG + +
12 Lysine utilization + +
13 Esculin hydrolysis + +
14 Arginine utilization + +
15 Lactose - -
16 Xylose - -
17 Maltose + -
18 Fructose + +
19 Dextrose + +
20 Galactose + +
21 Raf nose - -
22 Trehalose + +
23 Melibiose - -
24 Sucrose + +
25 L-Arabinose + +
26 Mannose + +
27 Inulin + +/-
28 Sodium gluconate + +
29 Glycerol + +
30 Salicin + +
31 Dulcitol + +
32 Inositol + +
33 Sorbitol + +/-
34 Mannitol + +
35 Adonitol - +
36 Arabitol - +
37 Erythritol + +
38 alpha-Methyl-D-glucoside + +
39 Rhamnose + -
40 Cellobiose + +
41 Melezitose - +
42 alpha-Methyl-D-Mannoside + -
43 Xylitol - +
44 ONPG - +
45 Esculin - +
46 D-Arabinose + +
47 Citrate utiliation - +
48 Malonate - -
49 Sorbose + +
Gayatriben B. Patel and Kamlesh R. Shah
510 BIODEGRADATION OF COTTON SEED SOAPSTOCKS BY NOVEL INDIGENOUS
BACILLUS SP.
BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS
lowed by Pseudomonas spp. 18%. Bacillus sp. has been
potential for production of proteases and lipases (Sangee-
tha et al., 2010). Bacterial Bacillus species are the promi-
nent source of lipases in which B. subtilis (Shah et al.,
2006), Bacillus pumilus (Sangeetha et al., 2008), Bacillus
licheniformis (Madan et al., 2009, Sangeetha et al., 2010)
are potent strains for lipase enzyme production. For the
enzyme production we have done plate assay with vari-
ous enzymes like lipase, cellulase, protease from which
B.pumulis gives maximum zone of hydrolysis of 1.37
mm with lipase and 3.10mm with protease while Bacil-
lus licheniformis and Bacillus pumulis gives same maxi-
mum zone of hydrolysis of 1.80 mm with cellulase. From
the above isolated organisms, Bacillus pumilus, Bacil-
lus licheniformis isolates were found to be true lipase,
protease and cellulase producers giving maximum zone
of hydrolysis. Further screening was done for various
enzymes. The isolate 3B and 18B which further quan-
titatively tested for production of crude lipase by pNPP
as assay substrate and are ef cient to produce 1.72 U/
ml and 2.59 U/ml of crude lipase enzyme respectively
depicted in gure 2.
In Statistical Analysis p- value analyzes for control,
3B and 18B lipase productions were <0.0016, <0.024 and
<0.020 respectively. Bacillus pumilus is the best possible
isolate having highest lipase production and was fur-
ther screened and optimized for lipase production. The
maximum amount of lipase production was obtained on
the day 2nd with recoverable enzyme activity gradu-
ally decreasing thereafter in shaking conditions. Con-
sequently, further studies were carried out on cultures
incubated for 3 days to obtain enzyme production. Bio-
mass production remained stable, after which the cul-
ture reached the stationary phase. This forces microbes
to produce enzymes to degrade crude oil to utilize it as
a source of energy and these enzymes which were capa-
ble of producing certain secondary metabolites (Guru et
al., 2013, Adnan et al., 2018) P. gessardi was a novel
lipase degrading bacteria from the oil spilled soil which
can be useful for the remediation of oil contaminated
soil. (Veerabagu et al., 2014), Pseudomonas synxantha
PS1 a lipase producing strain from oil well produced
water having strong biodegradabitility of waste grease
(Cai et al., 2016) as well as fungi aspergillus nigar able
to degrade petroleum hydrocarbon (Mauti et al., 2016)
It can be concluded that from the results of the present
study that Bacillus pumilus could be used as new potent
microbial source of lipase. In further studies pilot scale
production and puri cation studies will be conducted.
CHARACTERIZATION OF BACTERIAL ISOLATES
The bacterial isolates which showed maximum zone of
clearance for lipase production were subjected to further
characterization and identi cation by morphological,
biochemical and molecular (by 16s rDNA sequencing)
characteristics. The culture code 3B known as Bacillus
licheniformis with accession Number KU728636 and
18B known as known as Bacillus pumiluswith acces-
sion Number KU728634. Phylogenetic trees are show in
Fig. 3 and 4.
From the table-2 we conclude from microscopic, mor-
phological, cultural characteristics and biochemical studies
that the organism is gram positive rod, aerobic, mesophilic,
highly mobile, non-endospore former, lactose non ferment-
ing Bacillussp..Based on its morphological and physiologi-
cal characteristics, the isolates were given for 16s r RNA
and it was con rmed that they belong to Bacillus genus.
CONCLUSION
Screening for lipase producing cultures from cotton oil
re nery industries and resulted in the isolation of 49
isolates including bacteria & fungi.The isolate which
showed highest production of lipase in plate assays were
further quantitatively tested for production of lipase by
pNPP as substrate assay. The culture was identi ed by
morphological and molecular basis as Bacillus licheni-
formis & Bacillus pumilus. Culture was deposited in the
NCBI culture collection center with accession number.
Presence of cellulase and protease enzyme may help in
degradation study.
ACKNOWLEDGMENTS
The authors wish to express their thanks to the Depart-
ment of Biotechnology, Pramukh Swami Science and
H.D. Patel Arts College, Kadi. Gujarat State Biotechnol-
ogy Mission at Gandhinagar for the valuable advices in
the identi cation of the Bacterial Spp.
REFERENCES
Adnan B. Al- Hawash, Maytham A. Dragh, Shue Li, Ahmad
Alhujaily, Hayder A., Xiaoyu Zhang and Fuying Ma (2018):
Principles of microbial degradation of petroleum hydrocarbons
in the environments. Egyptian journal of Aquatic Research.;
44:71-76.
Ali L., Nangyal H., Wali A., Gul-e- Sahra, Ahmad T. (2014):
Screening of oil contaminated soil for isolation of lipids
degrading bacteria. Sci. Int. (Lahore); 26: 1595-1600.
Bharathi, D., G. Rajalakshmi, S. Komathi (2018): Optimization
and production of lipase enzyme from bacterial strains isolated
from potrol spilled soil. Journal of king saud university Sci-
ence; 1-4
Cai Xianghai, Siqi Chen, Hong Yang, Wel Wang, Lin Lin. Yal-
ing Shen, Wei Wei and Dong-ZhiWei (2016): Biodegradation of
waste grease and biochemical properties of a novel lipase from

Gayatriben B. Patel and Kamlesh R. Shah
BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS BIODEGRADATION OF COTTON SEED SOAPSTOCKS BY NOVEL INDIGENOUS
BACILLUS SP.
511
Pseudomonas synxantha PS1, Canadian journal of Microbiol-
ogy, 62: 588- 599.
Cappuccino, JGS N.J.G. Cappuccino, N. Sherman (1996):
Microbiology: A laboratory Manual (No. QR63. C36 1996).
Dabhi, BK Vyas R.V. and Shelat H.N. (2014): Use of Banana
Waste for the Production of Cellulolytic Enzymes under Solid
Substrate Fermentation Using Bacterial Consortium. Int. J.
Curr.Microbiol. App. Sci.; 3:337-346.
Guru G. S, Gohel H. R, Panchal M. R, Ghosh S. K and Braganza
V. B (2013): Isolation and Enrichment of Microbes for Degrada-
tion of Crude Oil. International Journal of Engineering Science
and Innovative Technology (IJESIT); 2:144-147.
Hanan S. A. (2012): Isolation and screening of extracellular
proteases produced by new Isolated Bacillus sp. Journal of
Applied Pharmaceutical Science; 2:071-074.
Hasan F., Shah A. A., and Hameed A. (2006): Industrial applica-
tions of microbial lipases. Enzyme Microb Techno; 39(2):235-
251.
King J. W., Taylor S., Snyder J. M., and Holliday R. (1998):
Total Fatty Acid Analysis of Vegetable Oil Soapstocks by
Supercritical Fluid Extraction/Reaction. JAOCS; 75:1291-l295.
Lokre S.S. and Kadam D.G. (2014): Screening of thermosta-
ble lipase producers from alkaline lake. Int. J. Curr. Microbiol.
App. Sci.; 3:240-245.
Madan, B., P. Mishra (2009): Over expression, puri cation and
characterization of organic solvent stable lipase from Bacillus
licheniformis RSP-09. Journal of molecular microbiology and
biotechnology; 17(3):118-123.
Mauti, GO., J. Onguso, D. K. Kowanga and E. M. Mauti (2016):
Biodegradation activity of Aspergillus niger Lipase isolates
from a tropical country Garage. Journal of Scienti c and Inno-
vative Research; 5(1):15-18,
Patagundi B. I., Shivasharan C. T. and Kaliwal B. B. (2014):
Isolation and Characterization of Cellulase producing bacteria
from Soil. Int. J. Curr. Microbiol. App. Sci; 3:59-69.
Prabavathi R., Mathivanan V. and Ambika A. (2012): Screening
of protease enzyme By construction of Metagenomic library
from Marine soil sediments. International Journal of Pharma
Sciences and Research; 3:396-399.
Pualsa J., Verma D., Gavankar R., and Bhagat R. D. (2013): Pro-
duction of microbial lipases isolated from Curd using waste oil
as a substrate. Research Journal of Pharmaceutical, Biological
and Chemical Sciences; 4:831- 839.
Sangeetha R., Geetha A., Arulpandi I. (2008): Optimization
of protease and lipase production by Bacillus pumilus SG 2
isolated from an industrial ef uents. The Internet Journal of
Microbiology; 5:61-65.
Sangeetha R., Geetha A., Arulpandi I. (2010): Concomitant pro-
duction of protease and lipase by Bacillus licheniformis vsg1:
Production, puri cation and characterization. Brazilian jour-
nal of microbiology; 41:179-185.
Saraswat, R V. Verma, S. Sistla and I. Bhushan (2017): Evalu-
ation of alkali and thermotolerant lipase from an indigenous
isolated Bacillus strain for detergent formulation. electronic
journal of biotechnology; 30:33-38.
Saxena R. K., Ghosh P. K., Gupta R., Davidson W. S., Bradoo S.
and Gulato R. (1999): Microbial lipases : Potential biocatalyst
for the future industry. Current Science; 77: 101-115.
Shah K. R., Patel P. M., Bhatt S. A. (2007): Lipase production by
bacillus sp. Under different Physio-chemical conditions.Jour-
nal of Cell and Tissue Research; 7:913-916.
Sharma R., Chisti Y. , Banerjee U. C. (2001): Production, puri-
cation, characterization, and applications of lipases. Biotech-
nology Advances; 19:627 – 662.
Sirisha E., Rajasekar N. and Narasu M. L. (2010): Isolation and
Optimization of Lipase Producing Bacteria from Oil Contami-
nated Soils. Advances in Biological Research; 4:249-252.
Vandana P., Peter J. K. (2014): Comparative Study for Lipase
Production by Using Pseudomonas aeruginosa and Pseu-
domonas uorescens. International journal of engineering sci-
ences & research technology; 3:31-34.
Veerapagu M., Sankara N. A., Jeya K.R., Alagendran S. (2014):
Isolation and Identi cation of a Novel Lipase Producing Bac-
teria from Oil Spilled Soil. International Journal of Innovative
Research in Science, Engineering and Technology; 3:18122
-18129.
Verma N., Thakur S. and Bhatt A. K. (2012) Microbial Lipases:
Industrial Applications and Properties. International Research
Journal of Biological Sciences; 1:88-92.
Winkler U. K., Stuckman M. (1997): Glycogen, hyluronate and
some other polysaccharides greatly enhance the formation of
exo-lipase by Serrati amarescens. J. Bacteriol; 138:663-670.