
Agricultural
Communication
Biosci. Biotech. Res. Comm. 11(2): 216-223 (2018)
Relationship among combining ability, heterosis and
genetic distance in maize (
Zea maize
L.) inbred lines
under water-de cit conditions using line × tester and
molecular analysis
Sharareh Fareghi
1
, Aghafakhr Mirlohi
2
* and Ghodratollah Saeidi
3
1
Student of Plant Breeding, Department of Agronomy and Plant Breeding, College of Agriculture, Isfahan
University of Technology, Isfahan, Iran
2,3
Professor of Genetics and Plant Breeding, Department of Agronomy and Plant Breeding, College of Agriculture,
Isfahan University of Technology, Isfahan, Iran
ABSTRACT
This study was conducted to investigate the relationships of combining ability, hybrid performance and genetic dis-
tance using Sequence Related Ampli ed Polymorphism (SRAP) data and other data sets obtained from analysis of
agronomic performance of the CIMMYT maize inbred lines. For this purpose, 13 lines and four testers were crossed
through controlled pollination in a line × tester design scheme to develop 52 hybrids. These hybrids were evaluated
together with two standard checks (KSC704 and KSC705) for grain yield under two soil moisture environments for
two years (2014-2015). Pair-wise genetic distance (GD) was estimated based on Jaccard (J) and simple matching (SM)
coef cients. The variance components of speci c combining ability (SCA) were higher than general combining ability
(GCA), hence non-additive gene effects contributed to hybrid performance. There was no coincidence between the
SRAP data and morphological assessments in this study. Signi cant and positive association of general combining
ability with mid parent heterosis (MPH) under drought stress conditions is an indicator that GCA can be useful to
predict MPH during selections under water stress conditions. However, correlations of genetic distances with heterosis
under both conditions were too low to be predictive of hybrid vigor.
KEY WORDS: WATER STRESS, GENETIC DISTANCE, HYBRID PERFORMANCE, LINE × TESTER, SEQUENCE RELATED AMPLIFIED
POLYMORPHISM
216
ARTICLE INFORMATION:
*Corresponding Author: mirlohi@cc.iut.ac.ir
Received 12
th
April, 2018
Accepted after revision 17
th
June, 2018
BBRC Print ISSN: 0974-6455
Online ISSN: 2321-4007 CODEN: USA BBRCBA
Thomson Reuters ISI ESC / Clarivate Analytics USA and
Crossref Indexed Journal
NAAS Journal Score 2018: 4.31 SJIF 2017: 4.196
© A Society of Science and Nature Publication, Bhopal India
2018. All rights reserved.
Online Contents Available at: http//www.bbrc.in/
DOI: 10.21786/bbrc/11.1/4

Sharareh, Aghafakhr and Ghodratollah
INTRODUCTION
Maize is one of the most important crops worldwide,
which serves as food, animal feed and raw materials of
bioenergy. It is stated that, maize is queen of cereal crops
due to high yielding potential and genetic diversity. The
global production of this crop has increased during last
years. However, its yield is reduced due to water de -
cit, which is one of the most important environmental
factors affecting agricultural productivity worldwide,
(Prasanna, 2012, Aminu et al., 2014 Li et al., 2017).
Heterosis, a powerful phenomenon in the evolution
of plants, has been used extensively in crop produc-
tion. However, identi cation and selection of appropri-
ate parental combinations which produce superior F1
hybrids, is one of the most important stages in hetero-
sis utilization (Mohammed et al., 2014). Hybrid breeders
have always been concerned to the selection of appro-
priate parental lines without making all of the possible
crossing among the available lines. Selection of parents
with various genetic backgrounds is hardly substantial
in the development of hybrids having optimal expres-
sion of heterosis (Hallauer et al., 2010 Pheirim et al.,
2017).
During the past decades, several procedures have
been utilized for prediction of heterosis, including per-
formance of parental lines, combining ability, genetic
diversity which determined using multivariate analysis
of morphological and agronomic traits and molecular
markers (Mohammadi et al., 2008). Selection based on
phenotypic traits is extremely in uenced by environ-
mental factors, and the presence of genotype × envi-
ronment interactions can hide the actual genetic value.
Moreover, because of strong dominance effects of genes
controlling maize yield, hybrid performance may not be
predicted from the performance of parental lines, reli-
ably. Furthermore, in breeding programs with a large
numbers of inbred lines, making and evaluation of all
of the possible crosses is not only expensive and bor-
ing, but also practically dif cult and time consuming
(Mohammadi et al., 2008).
In maize, several methods have been expanded for the
prediction of hybrid performance by means of genetic
markers (Frisch et al., 2010; Maenhout et al., 2010). Con-
sidering the cost and time which is required for eld
evaluation of hybrids, the utilization of genetic mark-
ers for identi cation of best heterotic combination of
parental lines can be a suitable alternative (Mohammed
et al., 2014). Molecular markers have been widely uti-
lized in breeding programs, as a tool for the selection of
the best parental lines of crosses; and as potential tools
for the prediction of the heterosis from a certain cross.
Parental genetic distance has been considered as a
feasible indicator for hybrid performance (Melchinger,
1999). Breeders are strongly interested to the prediction
of hybrid performance from parental genetic distance.
Because the preferable crosses could be identi ed by
means of genetic distance before eld evaluation of all
hybrid combinations. This can increase the ef ciency
of hybrid breeding programs, (Mohammed et al., 2014).
Estimated genetic distances can be related with hybrid
performance from eld experiments, and the extension
of molecular marker systems such as sequence related
ampli ed polymorphisms (SRAP) have considerably
amended the power of the genetic distance estimation
between genotypes.
Several researchers have used genetic distance to pre-
dict hybrid performance (Dhliwayo et al., 2009; Devi and
Singh, 2011; George et al., 2011); however, their results
were inconsistent with each other. Some researchers
reported a positive correlation between marker based
genetic distance and hybrid performance (Amorim et
al., 2006; Srdic et al., 2007; George et al., 2011), while
other researchers have reported no correlation between
these two phenomenon (Balestre et al., 2008; Dhliwayo
et al., 2009; Devi and Singh, 2011). Hence, the poten-
tial utilization of molecular markers in predicting the
amount of hybrid performance in maize needs more
research. Though signi cant associations were found
between hybrid performance and genetic diversity in
several investigations, the level of association varied
widely from one study to another. Moreover, the reli-
ability of molecular markers in estimating genetic dis-
tance depends on several factors such as the number of
markers, their mode of inheritance and uniform distribu-
tion across the genome (Hahn et al., 1995; Mohammadi
et al., 2008).
To the best of our knowledge, there is a little infor-
mation about the association of genetic divergence and
hybrid performance in CIMMYT maize inbred lines,
which asks for studies to determine genetic distance as
suitable predictor of hybrid performance in this germ-
plasm. It is also needed to examine combining ability
of parents as predictor of heterosis and F1 performance
comparing with genetic distance measured by molecu-
lar markers. Therefore, this study was conducted to 1)
investigate the possibility of predicting the hybrid per-
formance using SRAP data and other data sets acquired
from analysis of agronomic performance of the CIMMYT
maize inbred lines; 2) determine associations among
genetic distance of molecular markers in parents, het-
erosis, F1 performance, general combining ability (GCA)
and speci c combining ability (SCA) effects of parents
and crosses, and compare the strategies to determine
hybrid performance based on parental genetic distance
(GD), GCA and SCA for heterosis; and 3) evaluation of
coincidence between the SRAP data and morphological
data.
BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS RELATIONSHIP AMONG COMBINING ABILITY, HETEROSIS AND GENETIC DISTANCE 217
Sharareh, Aghafakhr and Ghodratollah
MATERIALS AND METHODS
PLANT MATERIALS AND EXPERIMENTAL SITE
The experiment was conducted during two years (2014-
2015) at the Research Farm of Agriculture and Natural
Resources Research Center, Kermanshah, Iran (longitude
of 47° 26’ E, latitude of 34° 8’ N and altitude of 1346
m) on a silty clay loam soil. The mean annual precipita-
tion and temperature are 538 mm and 12.2 °C for the
region, respectively. In this study, a set of 13 inbred lines
were selected and crossed through controlled pollination
with four temperate maize testers using a line × tester
matting design to produce 52 hybrid combinations. The
origin and pedigree of the lines and testers are given in
Table 1.
FIELD EXPERIMENT
In this experiment, 52 hybrids derived from line × tester
matting scheme along with two standard checks were
planted in the eld according to a randomized com-
plete block design (RCBD) with three replications, at
two moisture environments (normal and water stress).
Each plot was included 2 rows of 4 m long with an
inter-row spacing of 0.75 m and in-row plant spacing
of 18 cm. Under the normal and stress moisture envi-
ronments, plants were irrigated when 50% and 65% of
the total available soil water was depleted from the root
zone, respectively. Soil moisture was measured based on
standard gravimetric methods (Clarke Topp et al., 2008).
The irrigation was applied by using a basin irrigat ion
system. The amount of water for each irrigation treat-
ment was measured using a volumetric counter. Grain
yield per plot was recorded on ve randomly selected
plants per replication.
SRAP ANALYSIS OF INBRED LINES
Genetic characterization of all of the inbred lines and
testers was done using a set of 30 SRAP primer pairs.
Genomic DNA was extracted from fresh leaves of each
line or tester according to the method of Murray and
Thompson (1980). PCR reactions were performed in a
10µl reaction mix and ampli ed products were resolved
by using 6% polyacrylamide gel followed by silver
staining.
STATISTICAL ANALYSES
Analysis of variance (ANOVA) for each moisture envi-
ronment was conducted using the PROC GLM of SAS
(SAS Institute 2008). Genotypes were considered as xed
effects while years, moisture environments and replica-
tions were considered as random. The SAS program was
used for the line × tester analysis to compute the SCA
effects (Singh and Chaudhary, 1977). The GCA effects
of lines and testers, the SCA effect of crosses, and their
interactions with the year were estimated based on the
factorial mating design as follows:
Y
ijk
=
μ
+
g
i
+ g
j
+ s
ij
+ e
k
+ ge
ik
+ ge
jk
+ se
ijk
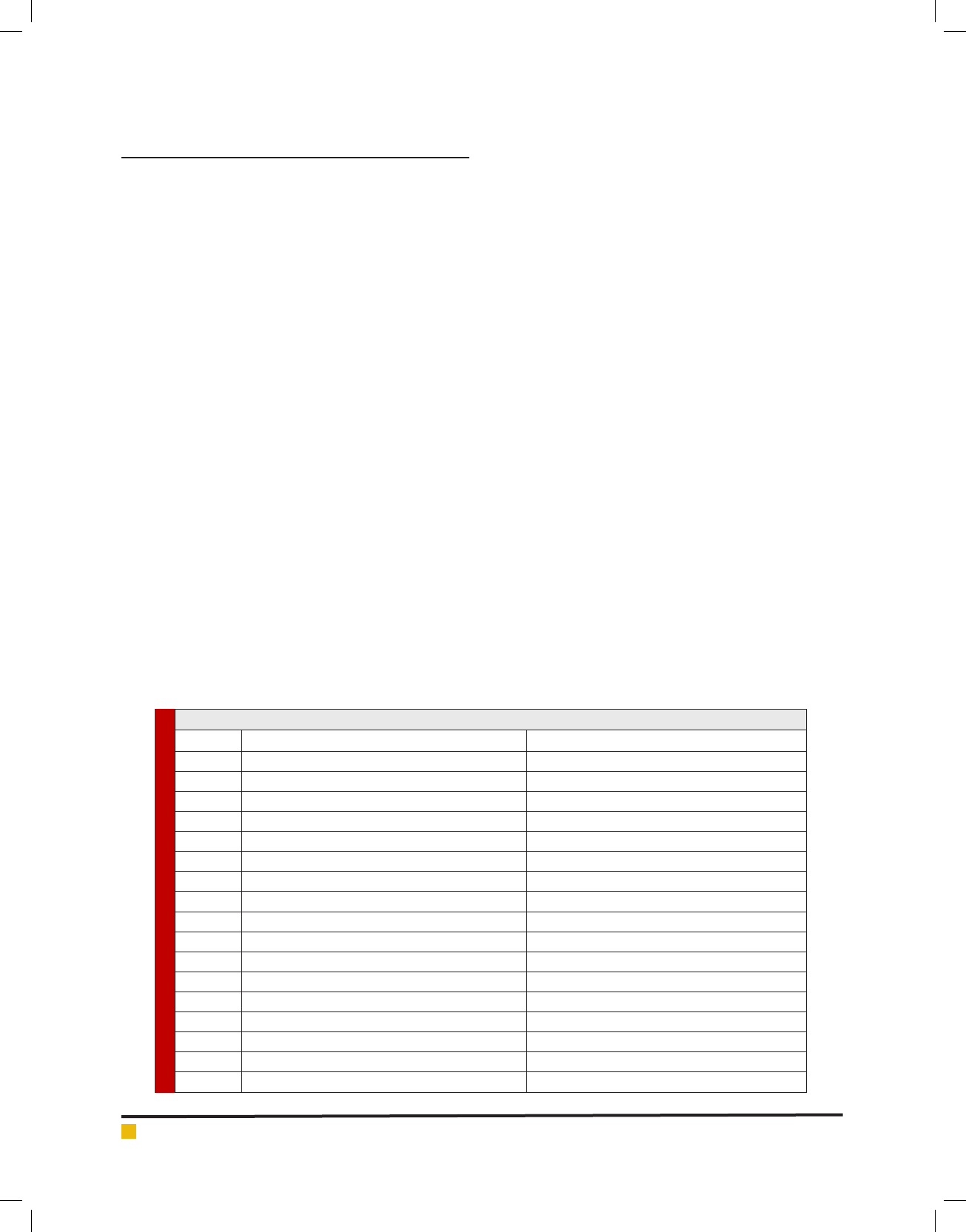
Table 1. Information on maize lines and testers used in the study
Parents Name of lines and testers/pedigree Origin
Line 1 4-CHTSEY,2002/1389/9=1390/13=1391/10 CIMMYT germplasm
Line 2 4-CHTSEY,2002/1389/19=1390/21=1991/70 CIMMYT germplasm
Line 3 7-CHTSEY,2002/1389/33=1390/33=1391/61 CIMMYT germplasm
Line 4 7-CHTSEY,2002/1389/35=1390/37=1391/64 CIMMYT germplasm
Line 5 K18 × 2-CHTHIY, 2002/1389/59=1390/73=1391/43 derived from cross k18 × CIMMYT originated line
Line 6 K18 × 2-CHTHIY, 2002/1389/61=1390/77=1391/46 derived from k18 × CIMMYT originated line
Line 7 XT03 Derived from unknown China -source
Line 8 4-CHTSEY, 2002/1390/5=1391/6 CIMMYT germplasm
Line 9 4-CHTSEY, 2002/1390/9=1391/8 CIMMYT germplasm
Line 10 7-CHTSEY, 2002/1390/41=1391/22 CIMMYT germplasm
Line 11 20-CHTSEY,2002/1390/45=1391/25 CIMMYT germplasm
Line 12 20-CHTSEY,2002/1390/53=1391/31 derived from CIMMYT germplasm
Line 13 MO17 × 6-CHTHEY, 2002/1390/69=1391/40 derived from cross MO17 × CIMMYT originated line
Tester 1 MO17 CL. 187–2 × C103
Tester 2 0K18 derived from MO17 changes
Tester 3 A679 A B73 back-cross derived line [(A662 × B73)(3)]
Tester 4 K166B derived from CIMMYT germplasm
218 RELATIONSHIP AMONG COMBINING ABILITY, HETEROSIS AND GENETIC DISTANCE BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS

Sharareh, Aghafakhr and Ghodratollah
Where Y
ijk
; is performance of the hybrid when ith line
is crossed to jth tester, in the kth year, is the overall
mean, g
i
is the effect of the ith line, g
j
is the effect of the
jth tester, s
ij
is the interaction of the ith line with the jth
tester, e
k
is the effect of the kth year, (ge)
ik
is the interac-
tion of the g
i
and e
k
, (ge)
jk
is the interaction of the g
j
and
e
k
, (se)
ijk
is the interaction of s
ij
and e
k
.
For each cross combination (P1 × P2) mid parent het-
erosis (MPH) was calculated according to Falconer and
Mackay (1996) as follows:
Mid parent heterosis (MPH) = [(F1- MP) / MP] × 100
where F1 is the mean of the F1 hybrid performance and
MP is mean performance of two parental inbred lines.
Better parent heterosis (BPH) was calculated as:
BPH = [(F1- BP) / BP] × 100
where BP = mean of the better parent.
Genetic distance (GD) between each pair of parents
was estimated from the binary matrix, using Jaccard and
simple matching coef cients through the NTSYSpc ver-
sion 2.0. Cluster analysis was done based on the UPGMA
method. For evaluation of the correlation between two
similarity matrices (molecular and phenotypic data),
Mantel test in NTSYS software was applied. Mean of the
trait in each moisture environment was used to calculate
correlation coef cients between genetic distance, grain
yield, MPH, BPH, GCA and SCA.
RESULTS AND DISCUSSION
Presence of an appropriate value of heterosis for grain
yield and predicting hybrid performance is important in
hybrid breeding programs. The degree of heterosis may
in uence by genetic diversity of the germplasm being
used. The magnitude of heterosis which was observed in
this study indicates that there is an opportunity to use
this germplasm for extending hybrid varieties appropri-
ate for stress and non-stress conditions.
Analysis of variance of grain yield for each of the
two moisture environments showed signi cant geno-
type effects, indicating the existence of genetic varia-
tion among the genotypes. However genotype × year
interaction was non-signi cant in both moisture envi-
ronments, indicating that the genotypes were consistent
over the years (Table 2). Nevertheless, signi cant dif-
ferences (p < 0.01) among parents and F1 hybrids in
both moisture environments were found and indicated
that the data was suitable for genetic analysis of line
× tester design. The mean squares for lines and testers
which determine the GCA effects were also signi cant
and showed the predominance of additive gene action
in controlling grain yield. However, the mean squares of
testers were higher than that of the lines in normal con-
ditions and it was vice versa for water stress conditions.
The signi cance of line × tester interaction revealed that
SCA was also important in the control of grain yield and
indicated that non-additive gene effects also play an
important role in the controlling of this trait (Table 2).
Average grain yield for parents (Table 3) and hybrid
combinations (Table 4) showed a remarkable reduction
under water stress and it ranged from 8.68 ton/ha for
L8 × T1 to 15.16 ton/ha for L9 × T4 under normal con-
ditions. However, under water stress conditions, this
ranged from 4.95 ton/ha for L10 × T1 to 11.99 ton/ha
for L5 × T3. The GCA effect was positive and signi -
cant for two parents of T3 and T4 under both normal
and water stress conditions (Table 3). However, under
normal conditions four parents and under water stress
conditions ve parents showed signi cant and positive
values of GCA for grain yield (Table 3). Under normal
conditions ve hybrids and under water stress condi-
tions seven hybrids expressed signi cant and positive
values of SCA for grain yield (Table 4). Moreover, under
normal conditions ve hybrids and under water stress
conditions eight hybrids showed signi cant and nega-
tive SCA for this trait.
Table 2. Analysis of variance (ANOVA) for
combining ability of total grain yield based on
line × tester matting design under normal and
drought stress conditions.
Source of variation df
Mean squares (MS)
NS S
Year (Y) 1 18.11 17.41
Block (R) 4 37.18** 1.20
Genotype (G) 53 14.99** 19.33**
F1 vs. Check 1 2.65 39.26
Check 1 35.02** 6.02*
F1 51 14.84* 19.20**
Lines (L) 12 21.31** 36.36**
Testers (T) 3 46.94** 35.45**
L×T 36 10.00** 11.89**
G×Y 53 2.56 0.85
(F1 vs. Check) × Y 1 0.65 5.38
Check × Y 1 1.84 0.04
F1 × Y 51 2.61 0.77
L × Y 12 2.85 0.38
T × Y 3 0.71 0.86
L × T × Y 36 2.69 0.95
Error 212 2.32 1.12
2
A - 1.96 1.91
2
D - 4.88 7.29
*,**Signi cant at 0.05 and 0.01 probability levels, respectively.
NS: Non-stress, S: Stress
BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS RELATIONSHIP AMONG COMBINING ABILITY, HETEROSIS AND GENETIC DISTANCE 219

Sharareh, Aghafakhr and Ghodratollah
Mid parent heterosis (MPH) and better parent het-
erosis (BPH) values were signi cant and positive for all
hybrids under both normal and water stress conditions.
MPH ranged from 70% for L10 × T1 to 230% for L7 × T3
under normal conditions and from 0.63% for L10 × T3
to 3.15% for L5 × T3 under water stress conditions. The
range of BPH was from 39% for L10 × T1 to 183% for L7
× T3 under normal conditions, and from 0.62% for L10
× T3 to 2.92% for L5 × T3 under water stress conditions
(Table 4).
SRAP data showed low genetic distances among
parental lines. Distances ranged from 0.181 for L12 ×
T4 to 0.423 for L11 × T2 based on Jaccard coef cient,
and from 0.142 for L12 × T4 to 0.358 for L9 × T1 based
on simple matching coef cient (Table 4). The average
genetic distance among inbred lines of this study based
on Jaccard and simple matching coef cients were 0.33
and 0.25, respectively; indicating low levels of poly-
morphism among them. Ndhlela et al. (2015) stated that
low genetic distances can be attributed to the mixing of
germplasm by CIMMYT for population improvement at
the expense of hybrid breeding.
In eld experiments, the most expensive and time
consuming step of hybrid breeding programs is the
identi cation of inbred lines expressing higher hetero-
sis (Mohammadi et al., 2008). Plant breeders often have
used SCA of hybrids in identifying better parental lines
for extension of hybrid combinations. However, when a
large numbers of inbred lines are available in a breeding
program, more useful tools are needed. In maize, genetic
distance determined by molecular markers is the main
strategy which has been followed for determination of
hybrid performance and its potential for this purpose has
been evaluated in several researches. In these researches,
the extent of correlation differed greatly from one trait
to another and also varied extensively with the germ-
plasm used in different studies.
The correlation coef cients of GD calculated based
on Jaccard and simple matching coef cients were neg-
ligible and non-signi cantly different from zero for
each of TGY, MPH and BPH (Table 5). Therefore, pre-
diction of hybrid performance for grain yield based on
genetic distance estimated by SRAP markers cannot be
a practical approach and this was in agreement with the
results of Dhliwayo et al. (2009), Devi and Singh (2011)
and Ndhlela et al. (2015). However, some studies have
reported powerful correlation between hybrid perfor-
mance and parental genetic distance (Melchinger, 1999;
Singh and Singh, 2004).
Mohammadi et al. (2008) suggested that insuf cient
genome coverage, sample size of the parental lines and
progenies and different levels of dominance effects on
traits are some important reasons for the low correla-
tion between genetic distance and hybrid performance.
Other possible reason for this issue is utilization of
unlinked markers to the trait in estimation of genetic
distance. For solving this problem, Bernardo (1992) sug-
gested identifying of speci c marker loci with close link-
age to chromosomal segments controlling target traits.
Although genetic distance was not a reliable predictor
of hybrid performance, some promising approaches such
as BLUP (Best Linear Unbiased Prediction) along with
molecular marker data have been extended for predict-
ing hybrid performance using genetic distances. How-
ever, in this study signi cant and positive associations
were observed between TGY, BPH and MPH with SCA
effects of crosses. Moreover, a signi cant and positive
correlation was found between GCA and MPH under
water stress conditions (Table 5). This correlation is an
indicator that GCA can be useful to predict MPH during
selections under water stress conditions.
In this study, correlations based on genetic distance
estimates using simple matching coef cient were relatively
higher than correlations based on GD estimated using Jac-
card coef cient. This shows that the extent of correlation
coef cient was not only impressed by the germplasm under
study, but also by the genetic distance measures.
Cluster analysis grouped the 17 lines and testers into
three major groups (Fig. 1). However, the cluster analy-
Table 3. Grain yield (GY) means and general combining
ability (GCA) values for parental lines used in this study
under normal (NS) and water stress (S) conditions.
Parents
Grain yield
(GY) (Ton/ha)
General combining
ability (GCA) (Ton/ha)
NS S NS
L1 5.85 2.77 -1.12
L2 4.14 3.07 0.41
L3 5.49 2.40 0.57
L4 6.39 3.85 0.98**
L5 5.30 2.72 0.37
L6 6.33 3.22 0.54
L7 3.77 2.73 0.07
L8 4.91 2.82 -1.90**
L9 4.17 2.62 0.65
L10 7.15 3.05 -0.83*
L11 6.18 3.21 -0.98**
L12 5.74 2.57 1.33**
L13 5.70 2.88 -0.09
T1 4.56 2.34 -0.71**
T2 4.80 2.71 -0.63**
T3 5.27 3.06 0.75**
T4 5.53 3.04 0.59**
*, ** Signi cant at 0.05 and 0.01 probability levels, respectively.
NS: Non-stress, S: Stress
220 RELATIONSHIP AMONG COMBINING ABILITY, HETEROSIS AND GENETIC DISTANCE BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS

Sharareh, Aghafakhr and Ghodratollah
Table 4. Grain yield (GY) means, speci c combining ability (SCA), mid parent heterosis (MPH) and better
parent heterosis (BPH) estimates for 52 F
1
hybrids of line × tester under normal (NS) and water stress (S)
conditions, and genetic distance (GD) between respective parental lines using simple matching (SM) and
Jaccard’s (J) coef cients based on SRAP markers.
Hybrids
GY (Ton/ha) SCA (Ton/ha) MPH (%) BPH (%)
GD J GD SM
NS S NS S NS S NS S
L1×T1 10.55 7.35 -0.01 -0.43 103** 188** 80** 166** 0.327 0.245
L1×T2 10.73 7.56 0.08 1.04* 101** 176** 83** 173** 0.359 0.250
L1×T3 12.59 8.24 0.56 -0.81 126** 183** 115** 170** 0.308 0.221
L1×T4 11.23 9.00 -0.63 0.20 97** 210** 92** 196** 0.350 0.275
L2×T1 12.32 6.87 0.23 -0.34 183** 154** 170** 124** 0.353 0.270
L2×T2 12.57 7.79 0.38 0.45 181** 170** 162** 154** 0.292 0.196
L2×T3 11.62 6.79 -1.94** -1.17* 147** 122** 121** 121** 0.290 0.206
L2×T4 14.72 10.92 1.32 1.06* 204** 257** 166** 255** 0.312 0.240
L3×T1 14.21 7.28 1.96** -0.28 183** 207** 159** 203** 0.276 0.211
L3×T2 12.21 7.63 -0.13 0.37 137** 199** 122** 182** 0.349 0.255
L3×T3 11.61 7.71 -2.11** -0.80 116** 183** 112** 152** 0.301 0.225
L3×T4 13.84 7.75 0.28 0.71 151** 185** 150** 155** 0.280 0.221
L4×T1 12.20 6.66 -0.46 -0.26 123** 115** 91** 73** 0.395 0.324
L4×T2 11.42 8.41 -1.33 -0.75 104** 157** 79** 119** 0.388 0.289
L4×T3 14.73 8.93 0.60 -0.39 153** 159** 130** 132** 0.350 0.270
L4×T4 15.15 10.00 1.19 1.40** 154** 191** 137** 160** 0.376 0.314
L5×T1 14.59 6.76 2.54** -0.11 196** 167** 175** 148** 0.388 0.324
L5×T2 10.82 8.36 -1.33 -1.56** 114** 208** 104** 207** 0.370 0.279
L5×T3 14.51 11.99 0.98 0.80 175** 315** 174** 292** 0.333 0.260
L5×T4 11.16 9.57 -2.20** 0.87 106** 232** 102** 215** 0.398 0.343
L6×T1 11.97 7.22 -0.26 -0.48 120** 160** 89** 124** 0.291 0.216
L6×T2 12.21 6.77 -0.10 -0.87 119** 129** 93** 111** 0.297 0.201
L6×T3 12.79 10.58 -0.90 -0.24 121** 237** 102** 229** 0.283 0.201
L6×T4 14.79 10.86 1.26 1.59** 149** 247** 134** 238** 0.306 0.235
L7×T1 11.78 6.47 0.03 0.92* 183** 155** 158** 137** 0.240 0.181
L7×T2 11.70 9.90 -0.14 -0.35 173** 264** 144** 262** 0.347 0.255
L7×T3 14.92 9.98 1.70* 0.40 230** 245** 183** 227** 0.331 0.255
L7×T4 11.46 10.74 -1.59* -0.96* 146** 272** 107** 254** 0.256 0.201
L8×T1 8.68 7.62 -1.10 0.43 83** 195** 77** 170** 0.232 0.176
L8×T2 10.11 5.09 0.24 -0.20 108** 84** 106** 80** 0.327 0.240
L8×T3 11.21 5.21 -0.04 0.91* 120** 77** 113** 71** 0.290 0.221
L8×T4 11.98 5.71 0.90 -1.14* 129** 95** 117** 88** 0.259 0.206
L9×T1 12.38 8.84 0.05 0.71 184** 256** 172** 237** 0.422 0.358
L9×T2 11.61 5.79 -0.81 -1.07* 159** 117** 142** 114** 0.408 0.314
L9×T3 13.02 7.16 -0.78 0.32 176** 152** 147** 134** 0.390 0.314
L9×T4 15.16 8.76 1.53* 0.04 212** 210** 174** 188** 0.412 0.358
L10×T1 9.92 4.95 -0.93 0.00 70** 84** 39** 62** 0.342 0.265
L10×T2 11.92 6.79 0.98 0.02 100** 136** 67** 123** 0.340 0.240
L10×T3 12.75 4.96 0.43 0.89 105** 63** 78** 62** 0.336 0.250
L10×T4 11.67 6.23 -0.48 -0.90* 84** 105** 63** 105** 0.354 0.284
L11×T1 11.07 5.42 0.38 -1.18** 106** 95** 79** 69** 0.373 0.324
L11×T2 11.59 10.48 0.80 1.82** 111** 254** 87** 227** 0.423 0.348
L11×T3 11.60 6.67 -0.56 0.81 103** 113** 88** 108** 0.368 0.309
L11×T4 11.39 5.82 -0.61 -1.45** 94** 86** 84** 81** 0.356 0.314
BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS RELATIONSHIP AMONG COMBINING ABILITY, HETEROSIS AND GENETIC DISTANCE 221

Sharareh, Aghafakhr and Ghodratollah
L12×T1 12.78 7.57 -0.24 0.34 148** 208** 123** 194** 0.220 0.172
L12×T2 13.20 7.82 0.09 0.40 150** 196** 130** 189** 0.385 0.304
L12×T3 14.88 8.88 0.39 0.06 170** 216** 159** 191** 0.298 0.235
L12×T4 14.07 8.62 -0.25 -0.80 150** 207** 145** 184** 0.181 0.142
L13×T1 9.41 5.26 -2.18** 0.69 83** 102** 65** 83** 0.329 0.240
L13×T2 12.93 6.58 1.25 0.72 146** 136** 127** 129** 0.234 0.147
L13×T3 14.72 6.06 1.66* -0.80 168** 104** 158** 98** 0.310 0.216
L13×T4 12.16 5.20 -0.73 -0.62 117** 76** 113** 71** 0.353 0.270
*, **Signi cant at 0.05 and 0.01 probability levels, respectively.
NS: Non-stress, S: Stress
Table 5. Correlation coef cients of grain yield (GY), Mid parent heterosis (MPH) and Better
parent heterosis (BPH) with each of SRAP-based genetic distance estimates using Jaccard
(GDJ) and simple matching (GDSM) coef cients, general and speci c combining ability (GCA
and SCA, respectively) under normal (NS)and water stress (S) conditions.
Environments
GY (Ton/ha) MPH (%) BPH (%)
NS S NS S NS S
GDJ -0.023 0.028 -0.028 -0.03 -0.063 -0.045
GDSM 0.027 0.062 0.006 0.012 -0.02 -0.015
GCA 0.012 -0.243 0.049 0.661** 0.202 -0.073
SCA 0.689** 0.323** 0.565** 0.318** 0.528** 0.309**
*, **Signi cant at 0.05 and 0.01 probability levels, respectively.
NS: Non-stress, S: Stress
GY, grain yield; MPH, Mid parent heterosis; BPH, Better parent heterosis; GDJ, Jaccard genetic distance; GDSM, Simple
matching genetic distance; GCA, General combining ability; SCA, Speci c combining ability.
sis based on phenotypic traits and SRAP markers could
not separate parents based on geographical or ecological
data. Moreover, in this study there was no coincidence
between the SRAP data and morphological estimations,
which indicated poor association and agreement of
molecular marker diversity with that of phenotypic one
(r= 0.15 under normal and r= 0.10 under water stress
conditions). Several reasons are given for the discord-
ance between these two sets of data. Accumulation of
some characteristics having adaptive value in speci c
habitats subjected to similar ecologic conditions (Steiner
and Los Santos, 2001), differences between the evolu-
tionary rates of phenotypic traits with adaptive value
and those originating from selectively neutral DNA (Lin-
hart and Grant, 1996), selection pressure for homogeni-
zation of different traits in parental germplasm and the
different genomic regions evaluated with both markers
(Amini et al., 2011), are some of these probable reasons.
In conclusion, prediction of heterosis is critical and
valuable in hybrid breeding programs. In this regard,
a potentially powerful approach is the application of
genetic distance speci ed by molecular markers. In this
study, associations between genetic distance estimates
(GDJ and GDSM) with heterosis effects were negligible
and non-signi cant. Thus, prediction of heterosis based
on genetic distances estimated by SRAP markers cannot
be a practical approach. Use of unlinked markers to the
target traits, insuf cient genome coverage, sample size
of the parental lines and progenies and different levels
of dominance effects on target traits are some of prob-
able reasons for the low correlations between genetic
distance and hybrid performance. On the other hand,
identifying of speci c marker loci with close linkage
to chromosomal segments controlling target traits and
application of statistical methods such as BLUP along
with molecular marker data are some of the solutions
for this problem. A signi cant and positive association
FIGURE 1. Dendrogram depict-
ing genetic relationships among
parental lines involved in line ×
tester analysis, based on SRAP
data using UPGMA method and
Jaccard’s coef cient.
222 RELATIONSHIP AMONG COMBINING ABILITY, HETEROSIS AND GENETIC DISTANCE BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS

Sharareh, Aghafakhr and Ghodratollah
among GCA and MPH under drought stress conditions
is an indicator that GCA can be useful to predict MPH
during selections under water stress conditions.
REFERENCES
Amini, F., A. Mirlohi, M.M. Majidi, S. Shojaiefar, R. Kolliker
(2011) Improved polycross breeding of tall fescue through
marker-based parental selection. Plant Breeding 130: 701-
707.
Aminu, D., S.G. Mohammed, B.G. Kabir (2014) Estimates of
combining ability and heterosis for yield and yield traits in
maize population (Zea mays L.), under water conditions in the
Northern Guinea and Sudan Savanna zones of Borno State,
Nigeria. International Journal of Agriculture Innovations and
Research 2 (5): 824-830.
Amorim, E.P., V.B.O. Amorim, J.B. Dos Santos, A.P. De Souza,
J.C. De Souza (2006) Genetic distance based on SSR and grain
yield of inter and intra population maize single cross hybrids.
Maydica 51: 507–513.
Balestre, M., R.G. Von Pinho, J.C. Souza, J.L. Lima (2008) Com-
parison of maize similarity and dissimilarity genetic coef -
cients based on microsatellite markers. Genetics and Molecular
Research 7: 695–705.
Bernardo, R (1992) Relationship between single-cross perfor-
mance and molecular marker heterozygosity. Theoretical and
Applied Genetics 83: 628-634.
Clarke Topp, C., G.W. Parkin, T.P.A. Ferre (2007) Soil water
content. In: M.R. Carter, E.G. Gregorich, editors, Soil sampling
and methods of analysis. Canadian Soc. Soil Sci., Pinawa.
Devi, P., N.K. Singh (2011) Heterosis, molecular diversity, combin-
ing ability and their inter-relationships in short duration maize
(Zea mays L.) across the environments. Euphytica 178: 71–81.
Dhliwayo, T., K. Pixley, A. Menkir, M. Warbuton (2009) Com-
bining ability, genetic distances and heterosis among elite
CIMMYT and IITA tropical maize inbred lines. Crop Science
49: 1201–1210.
Falconer, D.S., T.F.C. Mackay (1996) Introduction to quantita-
tive genetics (4th ed.). Longman, London.
Frisch, M., A. Thiemann, J. Fu, T.A. Schrag, S. Scholten, A.E.
Melchinger (2010) Transcriptome-based distance measures for
grouping of germplasm and prediction of hybrid performance
in maize. Theoretical and Applied Genetics 120: 441 - 450.
George, M.L.C., F. Salazar, M. Warbuton, L. Narro, F.A. Vallejo
(2011) Genetic distance and hybrid value in tropical maize
under stress and non-stress conditions in acid soils. Euphytica
178: 99–109.
Hahn, V., K. Blankenhorn, M. Schwall, A.E. Melchinger (1995)
Relationships among early European maize inbreds. III. Genetic
diversity revealed with RAPD markers and comparison with
RFLP and pedigree data. Maydica 40: 299-310.
Hallauer, A.R., M.J. Carena, J.B. Miranda Filho (2010) Hand-
book of plant breeding: quantitative genetics in maize breed-
ing. Springer, New York.
Li, H., Q. Yang, N. Fan, M. Zhang, H. Zhai, Zh. Ni, Y. Zhang (2017)
Quantitative trait locus analysis of heterosis for plant height and
ear height in an elite maize hybrid zhengdan 958 by design III.
BMC Genetics 18: 36. DOI 10.1186/s12863-017-0503-9.
Linhart, Y.B., M.C. Grant (1996) Evolutionary signi cance of
local genetic differentiation in plants. Annual Review of Ecol-
ogy and Systematics 27: 237–277.
Maenhout, S., B.D. Baets, G. Haesaert (2010) Prediction
of maize single-cross hybrid performance: support vector
machine regression versus best linear prediction. Theoretical
and Applied Genetics 120: 415 - 427.
Mantel, N. A (1967) The detection of disease clustering and
a generalized regression approach. Cancer Research 27: 209-
220.
Melchinger, A.E (1999) Genetic diversity and heterosis. In:
J.G. Coors, S. Pandey, editors. The Genetics and Exploitation
of Heterosis in Crops, 99-118. ASA, CSSA and SSA, Madison,
WI, USA.
Mohammadi, S.A., B.M. Prasanna, C. Sudan, N.N. Singh (2008)
SSR heterogenic patterns of maize parental lines and predic-
tion of hybrid performance. Biotechnoloy & Biotechnoloical
Equipment 22 (1): 541-547.
Mohammed, W., D.P. Pant, Ch. Mohan, M. Arif Khan (2014)
Predicting heterosis and F1 performance based on combing
ability and molecular genetic distance of parental lines in Ethi-
opian mustard. East African Journal of Sciences 8 (2): 105-124.
Murray, M.G., W.F. Thompson (1980) Rapid isolation of high
molecular weight plant DNA. Nucleic Acids Research 8: 4321-
4325.
Ndhlela, T., L. Herselman, K. Semagn, C. Magorokosho, C.
Mutimaamba, M.T. Labuschagne (2015) Relationships between
heterosis, genetic distances and speci c combining ability
among CIMMYT and Zimbabwe developed maize inbred lines
under stress and optimal conditions. Euphytica 204: 635–647.
Pheirim, R., R. Niyaria, P. Kumar Singh (2017) Heterosis predic-
tion through molecular markers. Rising 1 (1): 45-50.
Prasanna, B.M. (2012) Diversity in global maize germ plasm:
characterization and utilization. Journal of Biosciences 37(5):
843–855.
Rohlf, F.J. (1998) NTSYS-pc numerical taxonomy and multi-
variate analysis system. Version 2.0. Exeter Software, Setauket,
NY.
SAS Institute. 2008. ‘User’s guide. Release 9.2.’ SAS Inst. Inc.,
Cary, NC.
Singh, R.K., B.D. Chaudhary (1977) Biometrical methods in
quantitative genetic analysis. Kalyani Publishers, New Delhi.
Srdic, J., S. Miladenovic-Drinic, Z. Pajic, M. Filipovic (2007)
Characterization of maize inbred lines based on molecular
markers, heterosis and pedigree data. Genetika 39: 355–363.
Steiner, J.J., G.G. Los Santos (2001) Adaptive ecology of Lotus
corniculatus L. genotypes: I. Plant morphology and RAPD
maker characterizations. Crop Science 41: 552–563.
BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS RELATIONSHIP AMONG COMBINING ABILITY, HETEROSIS AND GENETIC DISTANCE 223